Early development and epigenetic regulation - Żylicz Group
Developmental processes are intricately governed by factors inherited from parents and the nourishing uterine environment. Recent breakthroughs have revealed the swift responsiveness of metabolism to nutrient fluctuations, functioning as a genuine signalling network.
In the laboratory of developmental epi-metabolism, our primary focus is on unravelling the role of metabolism and chromatin in orchestrating gene expression states.
Fundamental level
On the fundamental level, we want to elucidate the molecular mechanism of this regulatory interplay and its biological functions.
Translational level
On a more translational level, we strive to apply this mechanistic knowledge to address pressing issues in assisted reproductive technologies. Specifically, we aim to discern the factors contributing to early in vitro fertilization (IVF) failures, seeking potential strategies for enhancing embryo quality. By delving into the intricacies of how chromatin senses metabolic changes, we would like to shed light on how successful development is orchestrated.
Research projects within the laboratory of epi-metabolism are centered around three collaborative themes that aim to deepen our understanding of fundamental biological processes and improve assisted reproduction technologies:
1. Molecular mechanisms of how chromatin and metabolism regulate early mammalian development:
This theme focuses on unraveling the intricate molecular mechanisms through which chromatin and metabolism regulate gene expression during the critical phase of early mammalian development. The team explores fundamental questions such as how global changes in metabolism lead to specific outcomes, the causal relationship between chromatin alterations and gene expression changes during lineage specification, and the biological significance of the coupling between metabolism and epigenetics.
2. Role of environment in successful embryogenesis:
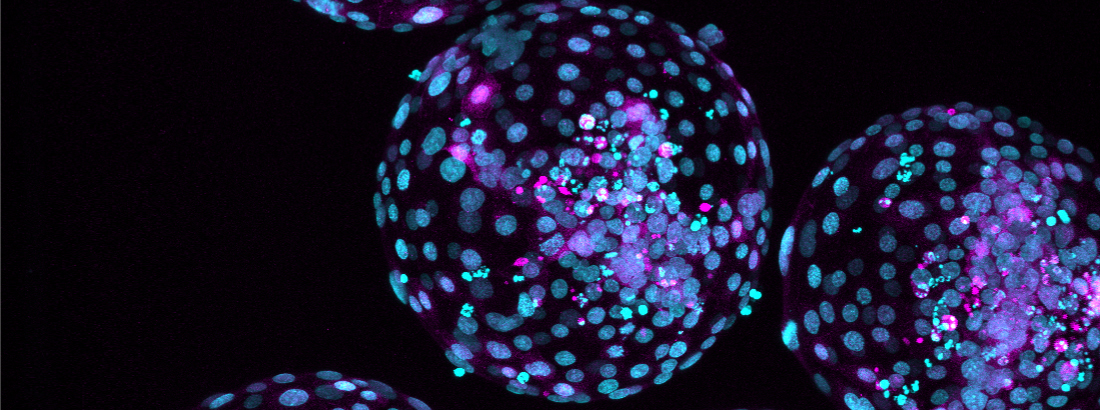
Within this theme, our researchers investigate the pivotal role of environmental factors, particularly nutrients, in the process of blastocyst formation and later at implantation. We delve into how stem cells perceive and respond to their surroundings, and how these cues translate into chromatin and gene expression programs that ultimately impact successful embryogenesis.
3. Advancements in assisted reproduction technologies:
Our most translational theme aims to leverage our molecular understanding of epi-metabolism and its interactions with environmental factors to develop improved media components for embryo culture during in vitro fertilization (IVF). By refining these technologies, we strive to enhance the success rates of assisted reproduction procedures.
To address the complexities of these research areas, we employ cutting-edge methodologies and experimental approaches. Our studies are grounded in the use of both mouse and human embryonic stem cells, as well as human blastoids, with a special emphasis on investigating the key supportive tissue known as the trophectoderm. Within these models, we employ a wide array of advanced tools, including metabolomics, epigenomics, and transcriptomics, to shed light on the intricate epi-metabolomic coupling that governs early development. To gain mechanistic insights our team complements these descriptive assays with functional CRISPR screens and precise degron manipulations. Such an integrative approach empowers us to uncover novel findings that contribute to the broader understanding of developmental biology and reproductive medicine.
List of publications by Jan Zylicz
Selected publications
- Stamidis, N., and Zylicz, J.J. + (2023). RNA-mediated heterochromatin formation at repetitive elements in mammals. EMBO J. 10.15252/embj.2022111717
- Pladevall-Morera, D., and Zylicz, J.J. + (2022). Chromatin as a sensor of metabolic changes during early development. Front Cell Dev Biol 10, 1014498. 10.3389/fcell.2022.1014498.
- Tjalsma, S.J.D. *, Hori, M. *, Sato, Y. *, Bousard, A., Ohi, A., Raposo, A.C., Roensch, J., Le Saux, A., Nogami, J., Maehara, K., Kujirai, T., Handa, T., Bages-Arnal, S., Ohkawa, Y., Kurumizaka, H., da Rocha, S.T., Zylicz, J.J. +, Kimura, H. +, and Heard, E+. (2021). H4K20me1 and H3K27me3 are concurrently loaded onto the inactive X chromosome but dispensable for inducing gene silencing. EMBO Rep. e51989. 10.15252/embr.202051989.
- Dossin, F., Pinheiro, I. *, Zylicz, J.J. *, Roensch, J., Collombet, S., Le Saux, A., Chelmicki, T., Attia, M., Kapoor, V., Zhan, Y., Dingli, F., Loew, D., Mercher, T., Dekker, J., and Heard, E.+ (2020). SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578, 455-460. 10.1038/s41586-020-1974-9.
- Zylicz, J.J. *, Bousard, A. *, Zumer, K., Dossin, F., Mohammad, E., da Rocha, S.T., Schwalb, B., Syx, L., Dingli, F., Loew, D., Cramer, P., and Heard, E. + (2019). The implication of early chromatin changes in x chromosome inactivation. Cell 176, 182-197 e123. 10.1016/j.cell.2018.11.041.
- Zylicz, J.J., Dietmann, S., Gunesdogan, U., Hackett, J.A., Cougot, D., Lee, C., and Surani, M.A. + (2015). Chromatin dynamics and the role of G9a in gene regulation and enhancer silencing during early mouse development. Elife 4. 10.7554/eLife.09571.
Hvidovre Hospital Fertility Clinic
The Developmental Epi-metabolic lab together with the team of Prof. Joshua Brickman is collaborating with the Hvidovre Hospital Fertility Clinic (Prof. Henriette Svarre Nielsen and Assoc. Prof. Nina la Cour Freiesleben) on a project seeking to identify key developmental bottlenecks during blastocyst development and how these could be overcome through epi-metabolic manipulations.
Lab of Prof. Thomas Moritz (NNF Center for Basic Metabolic Research)
The team also has an ongoing collaboration with the lab of Prof. Thomas Moritz (NNF Center for Basic Metabolic Research) on establishing high-sensitivity metabolomic approaches for developmental biology.
- ERC Starting Grant; "ChroMeta: Unravelling specificity of epi-metabolic regulation in mouse development" (2023-2028). Budget of 1,500,000 EUR.
- Lundbeck Foundation Fellowship; "Embryo and its environment: how metabolism shapes epigenome for development and life after birth?" (2021-2026). Budget of 1,342,282 EUR.
- Sapere Aude Starting Grant from Independent Research Fund Denmark; "Unravelling chromatin and metabolic regulatory networks during the first lineage choice of development" (2021-2025). Budget of 811,811 EUR.
- Project 1 Grant from Independent Research Fund Denmark; "Role of histone acetylation and DNA methylation in safeguarding development and stem cell differentiation" (2020-2023). Budget of 386,577 EUR.
Fellowships held by team members
- Lundbeck Foundation PostDoc Fellowship (2022-2025); Dr. Eleni Kafkia.
- Bridge Fellowship (2023-2025); Dr. Malene Hviid Saxtorph.

Żylicz Group is LEAF bronze certified on their sustainability actions.
Learn more about Zylicz Group
In the media
Associate Professor Jan Żylicz, was awarded a Lundbeck Foundation Fellowship for his project: 'Embryo and its environment: how metabolism shapes epigenome for development and life after birth?'
The movie is a production of Lundbeck Foundation.