Mechanics of tissue homeostasis - Sedzinski Group
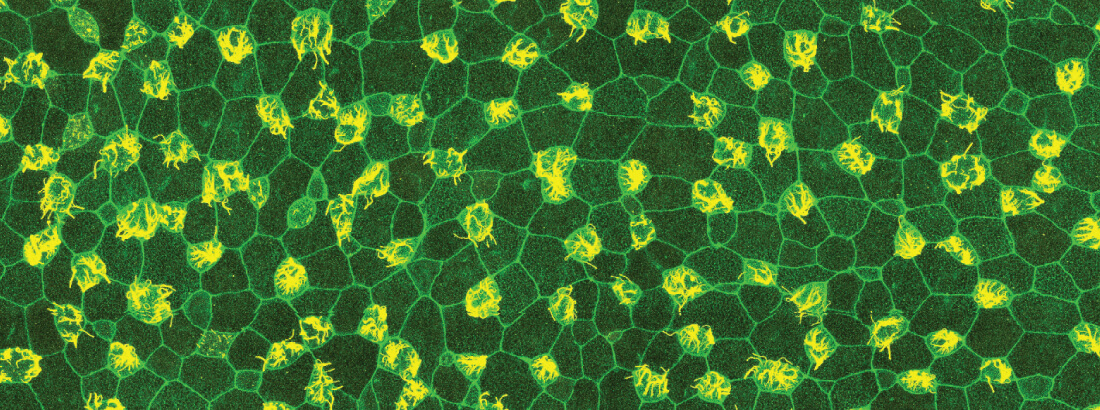
The research focus of the laboratory of Associate Professor Jakub Sedzinski at UCPH is to investigate the fundamental principles governing the development and regeneration of mucociliary epithelium (MCE). Specifically, they aim to elucidate the mechanical and molecular processes driving MCE differentiation and renewal from stem cells, with the ultimate aim of contributing to novel therapeutic approaches for respiratory diseases linked to impaired MCE.
To this aim, the group uses interdisciplinary methods, combining cell and developmental biology, physics and computation applied to developing frog embryos and human-derived respiratory organoids.
-
How tissue mechanics coordinate both cell fate and tissue morphogenesis
We want to understand the interplay between mechanical forces exerted by progenitor cell and generated within tissue to coordinate development of specialized tissues. -
Understanding temporal regulation of cell fate acquisition by mechanical forces
We want to determine when and how externally applied forces (stretch or compression) impact development of mucociliary epithelium. -
Integrating scRNAseq data with morphological parameters to generate a multimodal model of cell fate acquisition
We use computational methods to integrate morphological data describing cell shape changes and cell movement dynamics with scRNAseq data across the development of mucociliary epithelium. -
Establishing new models of human respiratory pathologies using 3D organoids
We use human-derived biopsies of respiratory tissues to engineer 3D organoids for studying respiratory diseases.
List of publications by Jakub Sedzinski
Selected publications
- Julie Lee, Andreas F.nss M.ller, Shinhyeok Chae, Alexandra Bussek, Tae Joo Park, Youni Kim, Hyun-Shik Lee, Tune H. Pers, Taejoon Kwon, Jakub Sedzinski* & Kedar Nath Natarajan (2023). A single cell time-resolved profiling of Xenopus mucociliary epithelium reveals non-hierarchical model of development. Science Advances. DOI: 10.1126/sciadv.add5745
- Guilherme Ventura, Aboutaleb Amiri, Raghavan Thiagarajan, Mari Tolonen, Amin Doostmohammadi, Jakub Sedzinski* (2022). Multiciliated cells use filopodia to probe tissue mechanics during epithelial integration in vivo. Nature Communications, doi: 10.1038/s41467-022-34165-0
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB (2017). RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. J Cell Science, doi: 10.1242/jcs.202234
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB (2016). Emergence of an apical epithelial cell surface in vivo. Developmental Cell, doi: 10.1016/j.devcel.2015.12.013
- Sedzinski J, Biro M, Oswald A, Tinevez JY, Salbreux G, Paluch E (2011). Polar actomyosin contractility destabilizes the position of the cytokinetic furrow. Nature, doi: 10.1038/nature10286
- Amin Doostmohammadi: theoretical modeling of morphogenesis / Niels Bohr Institute / University of Copenhagen, Denmark
- Aboutaleb Amiri: theoretical modeling of morphogenesis / Max Planck Institute for Physics of Complex Systems / Dresden, Germany
- Osvaldo Chara: theoretical modeling of morphogenesis / Faculty of Science / University of Nottingham, UK
- Elias Barriga: probing tissue mechanics / Gulbenkian Institute of Science (IGC) / Oeiras, Portugal
- Jonathan Brewer: probing tissue mechanics / South Denmark University / Odense, Denmark
- Poul Bendix: probing tissue mechanics / Niels Bohr Institute / University of Copenhagen, Denmark
- NNF Interdisciplinary Synergy Grant: Multiscale approach to engineer 3D bio-printed physiological human skin for use in basic and applied research.
- NNF Data Synergy Grant: MOPITAS – Multiomics profiling in time and space

Sedzinski Group is LEAF bronze certified on their sustainability actions.



