Epithelial homeostasis and regeneration - Jensen Group
Using primarily the intestinal epithelium as system to study cell behaviour, our goal is to understand how the epithelium is maintained long-term and how this is altered upon tissue damage to secure regeneration. We hope that such insights can be exploited to develop new therapies for ulcerating disorder.
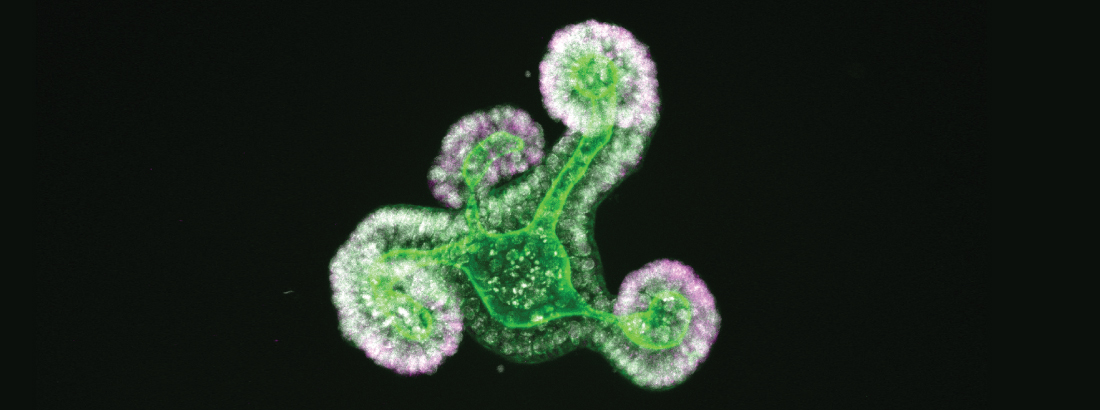
The epithelium of the intestine relies on stem cells for its constant and life-long replenishment. Recent evidence from our team has provided insight into how intestinal stem cells arise during late development from a more primitive fetal precursor to support maintenance of the homeostatic epithelium.
Interestingly, epithelial cells in the intestine adopt many of these primitive features during regeneration following damage. Moreover, the acquisition of these more primitive features appears to be a requirement for tissue regeneration.
In the Jensen group, we are interested in understanding the control mechanisms that ensure tissue maintenance during steady state homeostasis. This includes the correct or appropriate allocation of cells along the different intestinal lineages. Moreover, we are also interested in how these normal regulatory systems are bypassed following injury to fuel tissue regeneration.
Our long-term goal is to use knowledge from the analysis of in vitro organoid models, animal models of both homeostasis and regeneration, and patient samples to develop new therapeutic strategies for treating patients with diseases such as ulcerative colitis.
We have a number of ongoing projects in the team, which are driven either by individual scientists or as part of larger project groups. These can largely be divided into:
- Identify and characterise regulatory mechanisms controlling normal tissue homeostasis and lineage allocation.
- Functional characterisation of cell behaviour during tissue regeneration and probing regulatory mechanisms
- Development of treatment strategies for patients with ulcerative disorders
List of publications by Kim Bak Jensen
Selected publications
- Pikkupeura, L.M., Bressan, R.B., Guiu, J., Chen, Y., Maimets, M., Mayer, D., Schweiger, P.J., Hansen, S.L., Maciag, G.J., Larsen, H.L., Lõhmussaar, K., Pedersen, M.T., Teves, J.M.Y., Bornholdt, J., Benes, V., Sandelin, A.*, Jensen, K.B.* (2023) Transcriptional and epigenomic profiling identifies YAP signaling as a key regulator of intestinal epithelium maturation. Accepted Science Advances. DOI: 10.1126/sciadv.adf9460
- Hansen, S.L., Larsen, H.L., Pikkupeura, L.M., Maciag, G., Guiu, J., Müller, I., Clement, D.L., Mueller, C., Johansen, J.V., Helin, K., Lerdrup, M., Jensen, K.B.* (2023) An organoid-based CRISPR/Cas9 screen for regulators of intestinal epithelial maturation and cell fate. Accepted Science Advances. DOI: 10.1126/sciadv.adg4055
- Schweiger, P.J., Le Bouteiller, M., Yui, S., Thodberg, M., Clement, D., Jensen, K.B.* (2022) Extracellular matrix interactions provide tumor cells with an escape mechanism for commitment to differentiation. Gastroenterology 163:1688-90. DOI: 1053/j.gastro.2022.08.036
- Maimets, M., Pedersen, M.T., Guiu, J., Dreier, J., Thodberg, M., Antoku, Y., Schweiger, P.J., Rib, L., Bressan, R.B., Miao, Y., Garcia, K.C., Sandelin, A., Serup, P. and Jensen, K.B.* (2022) Mesenchymal-epithelial crosstalk shapes intestinal regionalisation via Wnt and Shh signalling. Nature Communications 13: 1-12. DOI: 1038/s41467-022-28369-7
- Guiu, J., Hannezo, E., Yui, S., Demharter, S., Ulyanchenko, S., Maimets, M., Jørgensen, A., Perlman, S., Lundvall, L., Mamsen, L.S., Larsen, A., Olesen, R.H., Andersen, C.Y., Thuesen, L.L., Hare, K.J., Pers, T.H., Khodosevich, K., Simons, B.D., Jensen, K.B.* (2019) Tracing the origin of adult intestinal stem cells. Nature 570: 107–111. DOI: 10.1038/s41586-019-1212-5
- Yui, S., Azzolin, L., Maimets, M., Pedersen, M.T., Fordham, R.P., Hansen, S.L., Larsen, H.L., Guiu, J., Alves, M.R.P, Rundsten, C.F., Johansen, J.V., Li, Y., Madsen, C.D., Nakamura, T., Watanabe, M., Nielsen, O.H., Schweiger, P.J., Piccolo, S.* and Jensen, K.B.* (2018) YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22:35-49. DOI: 1016/j.stem.2017.11.001
We collaborate with a number of research labs from across the world including a number of reNEW labs.
We receive support from a number of national and international funding schemes including Novo Nordisk Foundation, Danish Medical Research Council, Innovation Fund Denmark, European Union, and Neye Foundation.

Jensen Group is LEAF bronze certified on their sustainability actions.